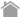Image based Protein-Protein interaction (PPI) analysis
- Detect PPI as fluorescent " Puncta " in living cells
- " Reversible " Puncta-formation
- " Easy -to-handle"
A New publication employing Fluoppi was just launched by Daiichi Sankyo RD Novare Co., Ltd.!
In this paper, Fluoppi technology was employed for spatiotemporal analysis of proteolysis-inducing regents such as PROTACs in the field of TPD.Kaji T, et al., Characterization of cereblon-dependent targeted protein degrader by visualizing the spatiotemporal ternary complex formation in cells. Sci Rep.10, Article Number: 3088 (2020) [PMID: 32080280]
A New publication about Fluoppi was jointly launched with A*STAR Laboratory!
In this paper, we demonstrated a multiplex Fluoppi assay analyzing p53-MDM2 and p53-MDM4 PPIs simultaneously. With this assay, stapled peptide-based drugs were successfully evaluated in living cells.Frosi Y, et al., Simultaneous measurement of p53:Mdm2 and p53:Mdm4 protein-protein interactions in whole cells using fluorescence labelled foci. Sci Rep. 9, Article Number: 17933 (2019) [PMID: 31784573]
A New publication describing how we invented Fluoppi was jointly launched with Dr. Atsushi Miyawaki from RIKEN!
In this paper, many potential applications of Fluoppi which can visualize Protein-Protein Interaction (PPI) in living cells are reported.Watanabe T, et al., Genetic visualization of protein interactions harnessing liquid phase transitions. Sci Rep. 7, Article number: 46380 (2017) [PMID: 28406179]
| PPI induction | PPI inhibition |
|
|
|
| mTOR(FRB) and FKBP12 | p53 and MDM2 |
>> watch the videos in large size
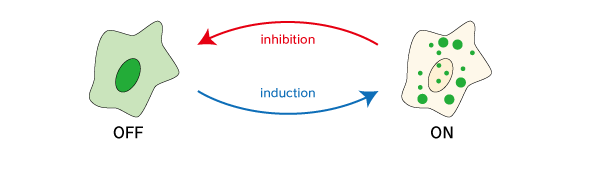
Fluoppi (Fluorescent based technology detecting Protein-Protein Interactions) is a novel technology to detect Protein-Protein Interaction (PPI) in living cells. Fluoppi detects PPI as fluorescent Puncta in reversible manner. The most important advantage of Fluoppi is that construction of the PPI detection system is very easy and does not need fine optimizations such like a linker sequence or length.
Key components
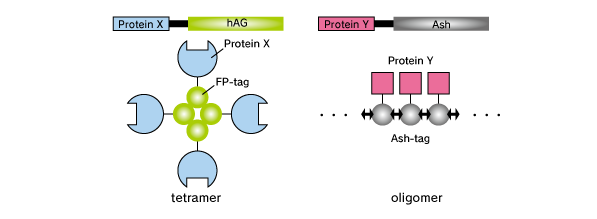
Fluoppi is a tag technology. Tetramer fluorescent protein (FP-tag) and Assembly helper tag (Ash-tag) are genetically fused to Protein X and Y, respectively. For the FP-tag, tetramer Green Fluorescent protein Azami Green (hAG) can be used. For the Ash-tag, PB1 domain of p62 can be used. PB1 is 102 amino acid length domain which has the capability of electrostatically homo-oligomerization in a head to tail manner (Saio et al. 2010). In our in vitro Fluorescence correlation spectroscopy analysis, Ash-tag behaved as tetramer to octomer in diluted solution.
Mechanism of action
Before protein X and Y interact, most of the molecule is disseminated.
When protein X and Y interact, these proteins associate each other to form detectable Puncta.
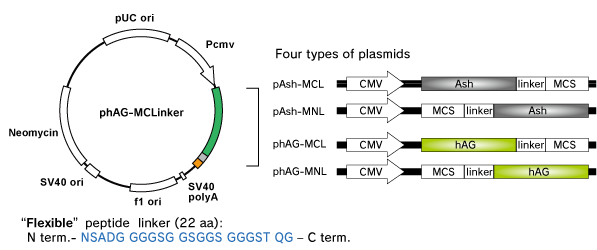
Fluoppi tags are able to work in both N and C terminal fusion. We have several plasmid vectors which include CMV promoter, Fluoppi tag, flexible peptide linker, Multiple Cloning Site (MCS) and Neomycin resistant gene.
Fluorescence characteristics
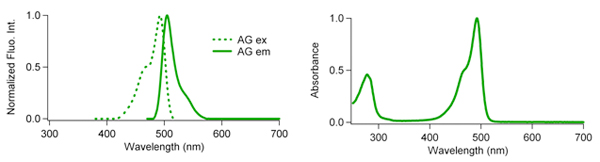
| Excit./Emiss.Maxima (nm) | Extinction coefficient (M-1cm-1) | Fluorescence quantum yield | pH sensitivity | |
| AG | 492/505 | 72,300 (492 nm) | 0.67 | pKa=5.0 |
1. Target gene amplification & restriction enzyme cut
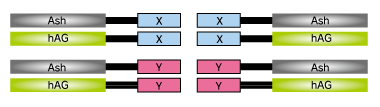
At first, proteins X & Y of your interest are fused to FP-tag and Ash-tag respectively.
We recommend to prepare all the eight possible constructs to identify the best workable combination.
2. Ligation & sequencing
3. Transfection
Co-transfect the pairs of plasmids.
4. imaging
Because fluorescent signal of Fluoppi is very high, conventional fluorescence microscopy can be used to image the cell.
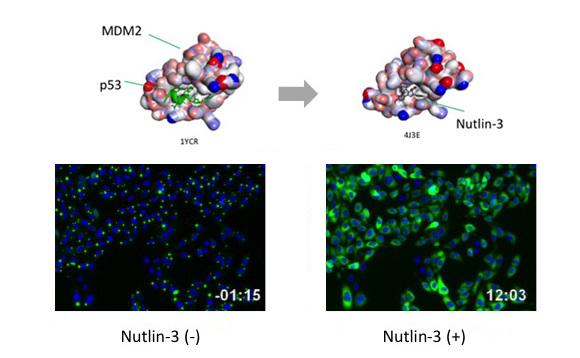
If the proteins interact with each other upon expression, fluorescent Puncta will be detected. Formation of Puncta is reversible so that they can be dissociated and the fluorescent signal will spread over the cell by PPI inhibitors, and vice versa by PPI inducer.
An example of p53-MDM2 PPI
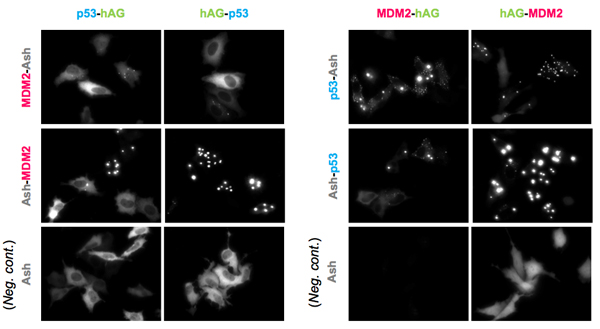
An example of p53-MDM2 PPI. (Partial sequence of p53 and MDM2 were used.)
Upper and middle 8 pictures represent 8 possible combinations. Lower 4 panels are results of negative control; hAG tagged protein and only Ash-tag were co-transfected. As shown in the result, Fluorescent patterns were different from combination to combination, so that we recommend to try all the 8 possible combinations to find the optimum combinations.
Localizations
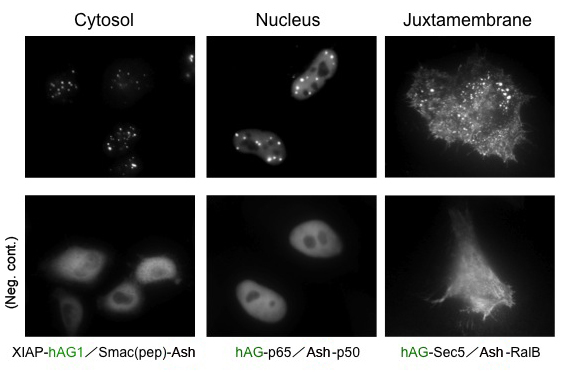
Because location of Puncta is not restricted to specific site inside the cell, Fluoppi can detect PPIs at several subcellular localizations such as cytosol, nucleus, and juxtamembrane. The upper pictures represent Puncta at several subcellular localizations, and the lower pictures are negative controls which express hAG tagged protein and Ash-tag without fusing any proteins. The images of juxtamembrane are taken by Total Internal Reflection Fluorescence Microscopy (TIRFM).
PPI modulator [induction]
mTOR (FRB domain) and FKBP12 is a well known PPI whose interaction is induced by Rapamycin. The pair of, mTOR(FRB)-hAG and Ash-FKBP12 was selected from eight possible combinations in a viewpoint of Puncta formation efficiency. After adding Rapamycin small Puncta were gradually formed in several minutes.
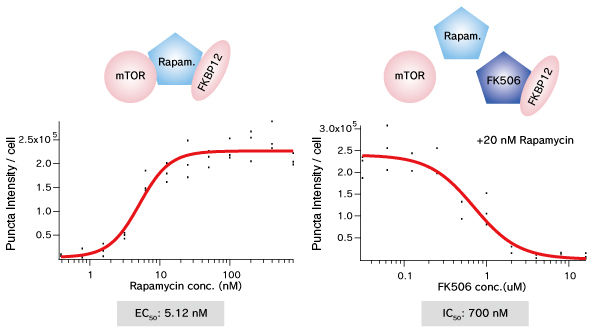
Dose dependent Puncta formations of Rapamycin (left) or competitive inhibition of Rapamycin by FK506 (right) were analyzed. Cells were treated with Rapamycin for 60 min in 37°C CO2 incubator followed by PFA fixation. Images were captured by ORCA-ER(Hamamatsu photonics) on IX71 fluorescence microscopy (Olympus). The spot detector plugin from ICY platform (Institut Pasteur) and IGOR software (HULINKS Inc.) were used for this analysis.
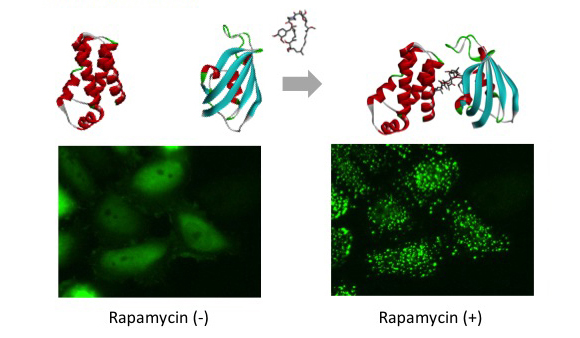
PPI modulator [inhibition]
p53-MDM2 is a famous target of PPI inhibitors in the field of anti cancer drug development. We applied this PPI to Fluoppi for demonstration. First, hAG-MDM2 and Ash-p53 was selected from the 8 pairs as represented above, then stable CHO-K1 cell line was established by using two selection marker; G418 and Hygromycin. Fluoppi plasmids including Hygromycin resistance gene is under released at present.
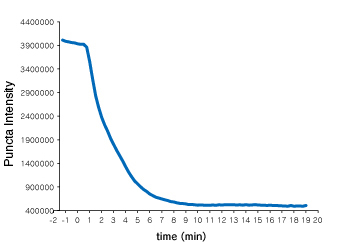 Time course of Puncta dissociation after adding Nutlin-3 was analyzed. Y axis represents fluorescent intensity of Puncta.
Time course of Puncta dissociation after adding Nutlin-3 was analyzed. Y axis represents fluorescent intensity of Puncta.
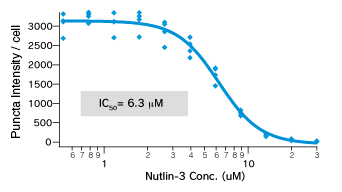 Dose dependent Puncta dissociations of Nutlin-3 were analyzed. Cells were treated with Nutlin-3 for 30 min followed by PFA fixation. Images were measured by InCellAnalyzer1000(GE healthcare).
Dose dependent Puncta dissociations of Nutlin-3 were analyzed. Cells were treated with Nutlin-3 for 30 min followed by PFA fixation. Images were measured by InCellAnalyzer1000(GE healthcare).
| Code No. | Product Name | DNA sequence |
|---|---|---|
| AM-8011M | Fluoppi Ver.2 : Ash-hAG (Ash-MNL/MCL + hAG-MNL/MCL) | pAsh-MNL Ver.2, pAsh-MCL, phAG-MNL, phAG-MCL |
| AM-8012M | Fluoppi Ver.2 : Ash-Red (Ash-MNL/MCL + Monti-Red-MNL/MCL) | pAsh-MNL Ver.2, pAsh-MCL, pMonti-Red-MNL, pMonti-Red-MCL |
| AM-8201M | Fluoppi : Ash-hAG [p53-MDM2] | pAsh/p53, phAG/MDM2 |
| AM-8202M | Fluoppi : Ash-hAG [mTOR-FKBP12] | pAsh/FKBP12, phAG/mTOR |
Publications
- Koyano F, et al., Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 510, 162-166 (2014) [PMID: 24784582]
- Yamano K, et al., Site-specific Interaction Mapping of Phosphorylated Ubiquitin to Uncover Parkin Activation. J Biol Chem. 290, 25199-211 (2015) [PMID: 26260794]
- Watanabe T, et al., Genetic visualization of protein interactions harnessing liquid phase transitions. Sci Rep. 7, Article number: 46380 (2017) [PMID: 28406179 ]
- Frosi Y, et al., Simultaneous measurement of p53:Mdm2 and p53:Mdm4 protein-protein interactions in whole cells using fluorescence labelled foci. Sci Rep. 9, Article Number: 17933 (2019) [PMID: 31784573]
- Kaji T, et al., Characterization of cereblon-dependent targeted protein degrader by visualizing the spatiotemporal ternary complex formation in cells. Sci Rep.10, Article Number: 3088 (2020) [PMID: 32080280]
References
- Saio T, et al., PCS-based structure determination of protein-protein complexes. J. Biomol. NMR. 46, 271-280 (2010) [PMID: 20300805]
- Banaszynski LA, et al., Characterization of the FKBP.rapamycin.FRB ternary complex. J. Am. Chem. Soc. 127, 4715-21. (2005) [PMID: 15796538]
- Vassilev LT, et al., In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 303, 844-848. (2004) [PMID: 14704432]
Please send us a completed and signed License Acknowledgement.
Please note that the product will be shipped after we receive a
completed and signed License Acknowledgement (PDF file on the right).
Prior to shipping, our staff may contact you to verify the intended use
of the product. Please also note that a separate agreement or a contract
may be required depending on the intended use.
The product you have ordered is sold for research purpose only. It is
not intended for industrial or clinical use, and shall not be used for
any purposes other than research. You are also asked not to hand over or
resell the product to a third party.
If you belong to a profit organization or a business corporation, or
wish to use the product for profit or commercial purposes, please
contact us.

- Fluorescent protein
- Basic fluorescent proteins
- Midoriishi-Cyan [Outline] [Product list]
- Umikinoko-Green [Outline] [Product list]
- Azami-Green [Outline] [Product list]
- Kusabira-Orange [Outline] [Product list]
- dKeima570 [Outline] [Product list]
- Keima-Red [Outline] [Detection of mitophagy with Keima-Red] [Product list]
- Photoconvertible fluorescent proteins
- Dronpa-Green [Outline] [Product list]
- Kaede [Outline] [Product list]
- Kikume Green-Red [Outline] [Product list]
- Organelle targeting vectors [Product list]
- Advanced fluorescent indicators
- Image based Protein-Protein interaction analysis "Fluoppi" [Outline] [Fluoppi Red] [Product list]
- Cell cycle indicator "Fucci" [Outline] [Product list]
- Protein-Protein Interaction Detection System "CoralHue™ Fluo-chase Kit" [Outline] [Product list]
- Anti-CoralHue™ fluorescent protein antibodies
- Anti-CoralHue™ fluorescent protein antibodies [Outline] [Product list]
- Resources
- CoralHue™ vectors full DNA sequence
- Spectrum