- Japan(Japanese / English)
- Global
- MBL TOP
- MBL site search
What is Selective autophagy?
Selective autophagy
Autophagy was initially thought to be a non-selective degradation mechanism, because the entire vesicle contents were digested. However, recent findings have revealed the selective degradation of mitochondria and other specific organelles, bacteria, and aggregates of proteins with attached ubiquitin chains (polyubiquitinated proteins). This mechanism is called “selective autophagy.”
Selective autophagy and p62
Because autophagosomes do not exhibit selectivity, “adaptor proteins” are necessary to link autophagosomes to proteins destined for selective degradation. One of these adaptor proteins is p62.
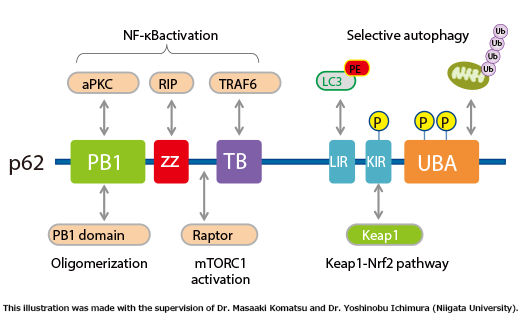
62/SQSTM1 is a scaffolding protein that interacts with various signaling molecules such as TRAF6, RIP, and aPKC (Fig. 1).
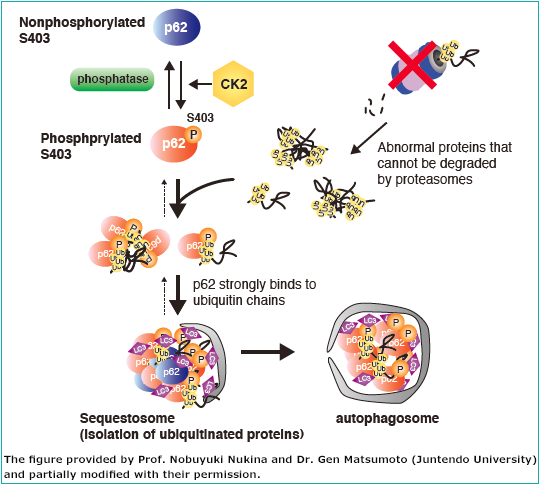
p62 contains an LC3-interacting region and is believed to be a substrate for selective autophagy. In addition, p62 contains a domain that binds ubiquitin chains, and mediates the recruitment of poly ubiquitinated protein aggregates and depolarized mitochondria to the autophagic machinery (see page 11 for the details of selective autophagy). In fact, in liver- and brain-specific autophagy-deficient mice, overaccumulation of p62 occurs, and ubiquitin- and p62-positive inclusion bodies are observed (Fig. 2). Importantly, ubiquitin- and p62-positive inclusion bodies are also observed in tissues of patients with neurodegenerative diseases (such as Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis), alcoholic hepatitis, hepatic steatosis, and liver cancer. There is increasing interest in the involvement of impaired autophagic degradation of p62 in these diseases.
■ p62/SQSTM1 domain structure (Fig. 1)
■ Selective autophagy of poly ubiquitinated protein by p62 (Fig. 2)



 p62 and phospho-p62 ELISA Kit
p62 and phospho-p62 ELISA Kit