- Japan(Japanese / English)
- Global
- MBL TOP
- MBL site search
Close
This site is for customers in Japan.
Customers in other regions, please go to Global page.
HOME > Product search results > Code No. PM040
Anti-Atg16L pAb
Price
¥41,800
Availability (in Japan)
10 or more
(In Japan at 00:05,
Jun 30, 2024 in JST)
Size
100 µL
| Data | |||||
|---|---|---|---|---|---|
| Clonality | Polyclonal | Clone | Polyclonal | ||
| Isotype (Immunized Animal) | Rabbit Ig (aff.) | ||||
| Applications |
|
||||
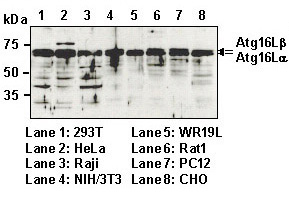
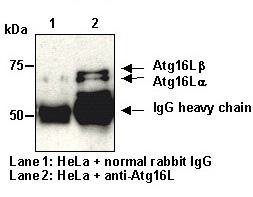
| Immunogen (Antigen) | Recombinant Human ATG16L1 TV2 (85-588 a.a.) | ||||
| Reactivity [Gene ID] | |||||
| Storage buffer | PBS/50% glycerol, pH 7.2 | ||||
| Storage temp. | -20°C | Conjugate | Unlabeled | Manufacturer | MBL |
| Alternative names | ATG16L1, autophagy related 16-like 1 (S. cerevisiae), IBD10, WDR30, APG16L, ATG16A, Apg16l | ||||
| Background | Autophagy is a process of intracellular bulk degradation in which cytoplasmic components including organelles are sequestered within double-membrane vesicles that deliver the contents to the lysosome/vacuole for degradation. Autophagy has two ubiquitin-like conjugation systems, the Atg12 and LC3-II systems. In the Atg12 conjugation system, the Atg16L-Atg12-Atg5 forms 800 kDa complex that elongate autophagic isolation membrane. After completion of the formation of the autophagosome, the Atg12-Atg5-Atg16L complex dissociates from the membrane. In recent study, nonsynonymous SNP analysis has indicated that ATG16L1 is a Crohn's disease susceptibility gene. | ||||
| Related products | 8485 Autophagy Ab Sampler Set M151-3 Anti-Atg10 (Human) mAb M154-3 Anti-Atg12 (Human) mAb M150-3 Anti-Atg16L mAb M153-3 Anti-Atg5 mAb PM050 Anti-Atg5 pAb PM036 Anti-LC3 pAb M152-3 Anti-LC3 mAb PD017 Anti-Beclin 1 pAb PM045 Anti-p62 (SQSTM1) pAb PM072 Anti-VMP1 pAb M133-3 Anti-Atg3 mAb PM039 Anti-Atg7 (Human) pAb |
||||
| Citations |
Western Blotting
Co-IP
Immunoprecipitation
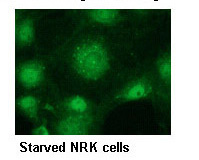
Immunocytochemistry
Immunofluorescence
Other(Image-based flow cytometry)
|
||||
| Product category |
|
||||
- The availability is based on the information in Japan at 00:05, Jun 30, 2024 in JST.
- The special price is shown in red color.
- Please note that products cannot be ordered from this website. To purchase the items listed in this website, please contact us or local distributers.
- Abbreviations for applications:
WB: Western Blotting, IH: Immunohistochemistry, IC: Immunocytochemistry, IP: Immunoprecipitation
FCM: Flow Cytometry, NT: Neutralization, IF: Immunofluorescence, RIP: RNP Immunoprecipitation
ChIP: Chromatin Immunoprecipitation, CoIP: Co-Immunoprecipitation
DB: Dot Blotting, NB: Northern Blotting, RNA FISH: RNA Fluorescence in situ hybridization - For applications and reactivity:
*: The use is reported in a research article (Not tested by MBL). Please check the data sheet for detailed information.
**: The use is reported from the licenser (Under evaluation or not tested by MBL).
- For storage temparature: RT: room temparature
- Please note that products in this website might be changed or discontinued without notification in advance for quality improvement.










 Citations
Citations Data Sheet
Data Sheet