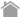- Japan(Japanese / English)
- Global
- MBL TOP
- MBL site search
RiboCluster Profiler™/RIP-Assay Kit, RIP-Certified Antibody
▼ A new article is published using a RiboCluster Profiler product.
RiboCluster Profiler™
- Biochemical approach to identify disease pathways and functionally related genes
- Tools for analysis of posttranscriptionally regulated and coordinated genes
- Useful tools to study disease genes and novel mechanisms of drug action
- Biochemical tools to improve microRNA target analysis


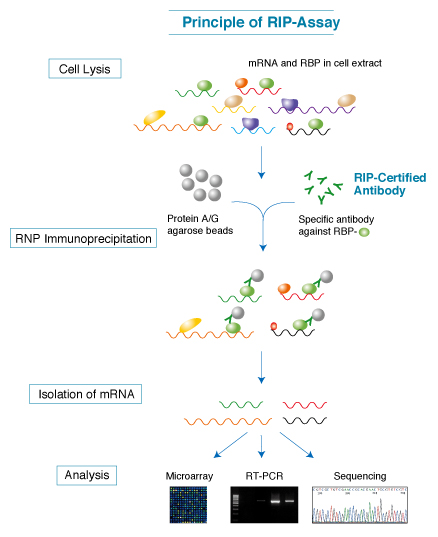
RIP-Chip technology
 MBL has developed and marketed the RIP-Assay Kit, which enables customers to immunoprecipitate the mRNA-RBP complexes with RBP specific antibodies. The RIP-Certified Antibodies against a large variety of RBPs are also available from MBL.
MBL has developed and marketed the RIP-Assay Kit, which enables customers to immunoprecipitate the mRNA-RBP complexes with RBP specific antibodies. The RIP-Certified Antibodies against a large variety of RBPs are also available from MBL.
RIP-Chip works on the same principle as the widely used ChIP-Chip. It immunoprecipitates the ribonucleoprotein (RNP) from the cell extracts using an antibody raised against the RBP of interest. This simple procedure is then followed by microarray analysis. While microarrays determine the sequences of the RNA targets by hybridization, direct sequencing approaches (RIP-Seq) can also be used to reveal RNA targets of RBPs.
RIP-Chip or RIP-Seq data provides insights into new cellular pathway components leading to potential therapeutic targets and can also provide informations regarding the effects of drugs on post-transcriptional processes. This technology can be applied to essentially any cellular system or animal model.
Ribonomic Discovery Cycle
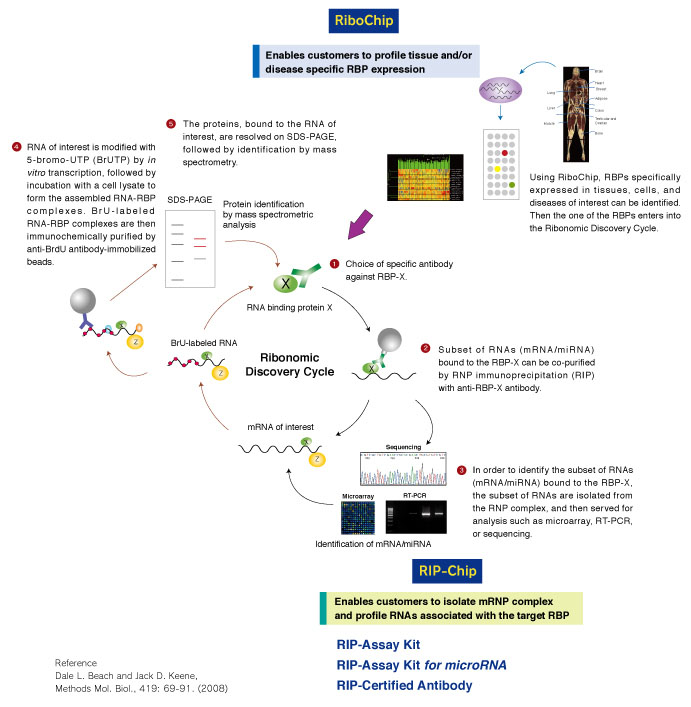
Example of analysis using RIP-Assay Kit
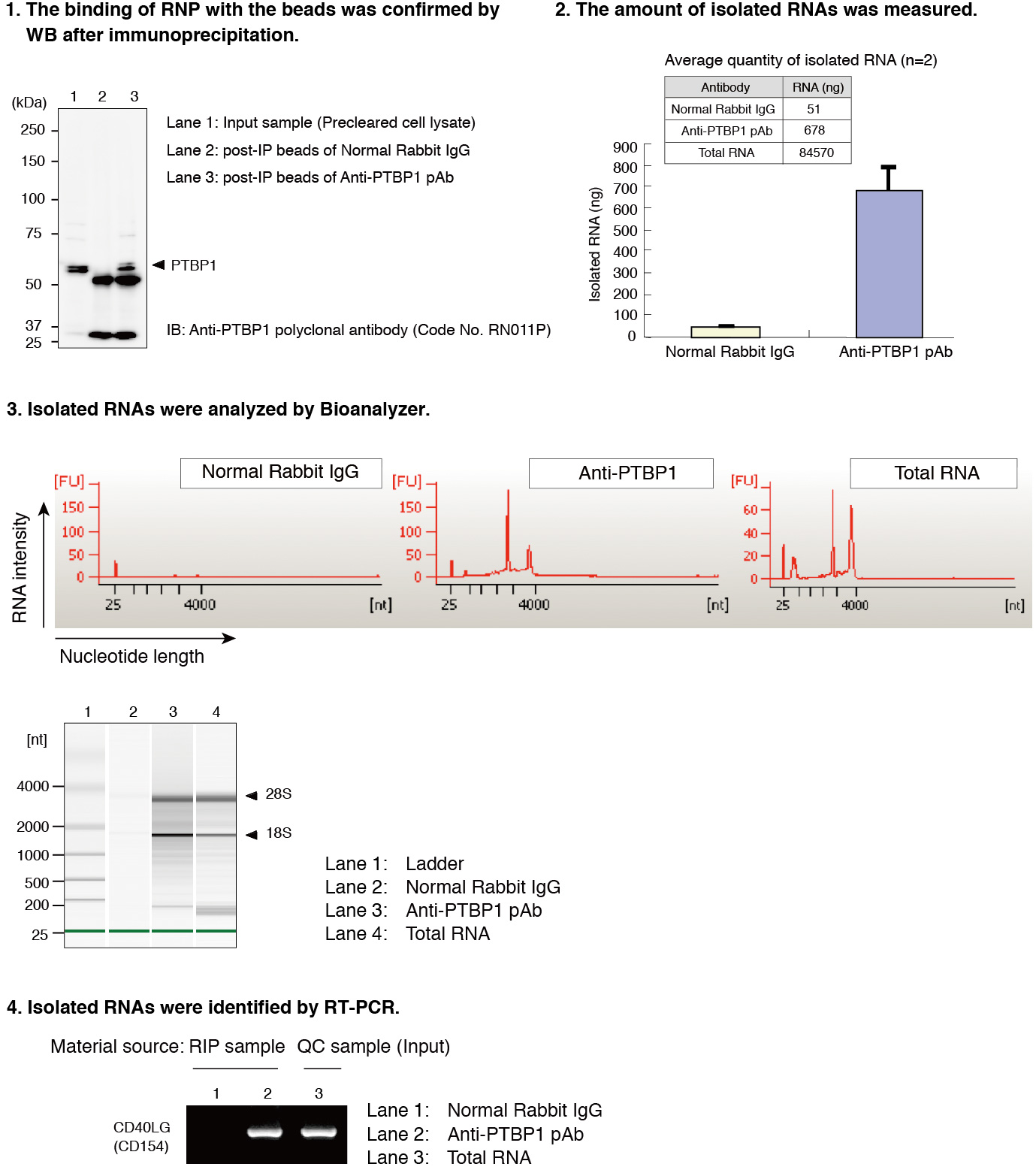
After the isolation of RNAs that bound with PTBP1 in Jurkat cells by using RIP-Assay Kit, the RNAs were identified by RT-PCR. CD40LG was detected at a significant level in the sample precipitated by anti-PTBP1 antibody rather than by control IgG. CD40LG has been reported to be target of PTBP1. Thus this data indicated the ability of RIP-Assay Kit for profiling the target mRNAs of RNP complex.
Publication list
- Janiszewska M, Suvà ML, Riggi N, Houtkooper RH, Auwerx J, Clément-Schatlo V, Radovanovic I, Rheinbay E, Provero P, Stamenkovic I., Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes & development 26, 1926-44 (2012) [PubMed: 22899010]
- Patel VL, Mitra S, Harris R, Buxbaum AR, Lionnet T, Brenowitz M, Girvin M, Levy M, Almo SC, Singer RH, Chao JA., Spatial arrangement of an RNA zipcode identifies mRNAs under post-transcriptional control. Genes & development 26, 43-53 (2012) [PubMed: 22215810]
- Lachke SA, Alkuraya FS, Kneeland SC, Ohn T, Aboukhalil A, Howell GR, Saadi I, Cavallesco R, Yue Y, Tsai AC, Nair KS, Cosma MI, Smith RS, Hodges E, Alfadhli SM, Al-Hajeri A, Shamseldin HE, Behbehani A, Hannon GJ, Bulyk ML, Drack AV, Anderson PJ, John SW, Maas RL., Mutations in the RNA granule component TDRD7 cause cataract and glaucoma. Science 331, 1571-6 (2011) [PubMed: 21436445]
- Jin D, Hidaka K, Shirai M, Morisaki T., RNA-binding motif protein 24 regulates myogenin expression and promotes myogenic differentiation. Genes Cells 15, 1158-67 (2010) [PubMed: 20977548]
- Thompson K, Victoria Lynn DiBona, Aditi Dubey, David Paul Crockett, Mladen-Roko Rasin, Acute adaptive responses of central sensorimotor neurons after spinal cord injury. Transl. Neurosci. 1, 268-278 (2010)
 If you are interested in miRNA analysis, learn about RIP-Assay Kit for microRNA
If you are interested in miRNA analysis, learn about RIP-Assay Kit for microRNA
Pick Up products

- RIP-Assay Kit [Outline] [Product]
- RIP-Assay Kit for microRNA [Outline] [Product]
- RIP-Certified Antibody [Products]
- RBP Antibody [Products]
- Isotype controls [Products]
- miRNA-mRNA paring analysis
- An emerging interest in RNA world
Introduction to post-transcriptional regulation of gene expression, function of non-coding RNAs and RNA-binding proteins