- Japan(Japanese / English)
- Global
- MBL TOP
- MBL site search
In Co-IP, complexes of two (or more) proteins are isolated using a procedure similar to the IP procedure.
Co-IP is often used for the analysis of interactions of multiple proteins and their functions.
The principle and method
The principle of Co-IP is the same as IP, except that the proteins associated with the antigen are also precipitated.
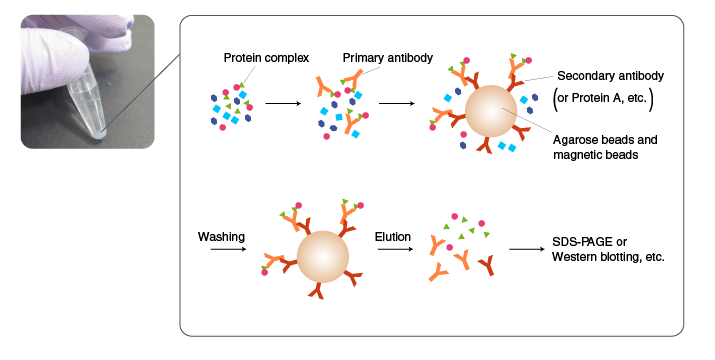
A protein complex is isolated by Co-IP using an antibody for one of the components in the complex.
The choice of antibody is critical for successful Co-IP. The antibody must bind to the surface of the complex. Alternatively, a component is tagged, and IP is performed using an anti-tag antibody. In either case, the method of cell lysis and the conditions of washing should be carefully evaluated because components in protein complexes are often held in place by weak interactions.
A common approach is to optimize the condition for Co-IP using a tagged protein, which is easy to detect, and then perform Co-IP experiments with endogenous proteins.
[Related topics] Pull-down assay experiments using Tagged protein purification kits
Complex formation identified by Co-IP should be confirmed by other methods, such as analysis of protein interactions in vivo using fluorescent-labeled proteins.
[Related topics] Protein-Protein Interaction analysis tool "Fluoppi"
[Related topics] Protein-Protein Interaction Detection System "CoralHue™ Fluo-chase Kit"

| Endogenous proteins | Tagged proteins (pull-down assay) | |
|---|---|---|
| Main advantages | Protein complexes are isolated in a relatively natural state. | An N- or C-terminal tag is likely available for antibody binding after complex formation. Antibody binding is unlikely to interfere with complex formation. |
| Issues to consider | The epitope may be buried upon complex formation. Antibody binding may interfere with complex formation. | The expression levels of recombinant proteins are substantially higher than those of their endogenous counterparts, which may result in artifactual results. |