| Citations |
Western Blotting - Saiki S et al. Caffeine induces apoptosis by enhancement of autophagy via PI3K/Akt/mTOR/p70S6K inhibition. Autophagy. 7, 176-87 (2011)(PMID:21081844)
- Ho H et al. WIPI1 coordinates melanogenic gene transcription and melanosome formation via TORC1 inhibition. J Biol Chem. 286, 12509-23 (2011)(PMID:21317285)
- Frankel LB et al. microRNA-101 is a potent inhibitor of autophagy. EMBO J. 30, 4628-41 (2011)(PMID:21915098)
- Starr T et al. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe. 11, 33-45 (2012)(PMID:22264511)
- Shi CS et al. Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 13, 255-63 (2012)(PMID:22286270)
- Morris CR et al. A knockout of the Tsg101 gene leads to decreased expression of ErbB receptor tyrosine kinases and induction of autophagy prior to cell death. PLoS One. 7, e34308 (2012)(PMID:22479596)
- Joubert PE et al. Chikungunya virus-induced autophagy delays caspase-dependent cell death. J Exp Med. 209,1029-47 (2012)(PMID:22508836)
- Murthy A et al. A Crohn's disease variant in Atg16l1 enhances its degradation by caspase 3. Nature 506, 456-62 (2014)(PMID:24553140)
- Silva BJ et al. Autophagy Is an Innate Mechanism Associated with Leprosy Polarization. PLoS Pathog. 13, e1006103 (2017)(PMID:28056107)
- Ye M et al. Curcumin Improves Palmitate-Induced Insulin Resistance in Human Umbilical Vein Endothelial Cells by Maintaining Proteostasis in Endoplasmic Reticulum. Front Pharmacol. 8, 148 (2017)(PMID:28377722)
- Kimura T et al. Age-dependent changes in synaptic plasticity enhance tau oligomerization in the mouse hippocampus. Acta Neuropathol Commun. 5, 67 (2017)(PMID:28874186)
- Takahashi A et al. Downregulation of cytoplasmic DNases is implicated in cytoplasmic DNA accumulation and SASP in senescent cells. Nat Commun. 9, 1249 (2018)(PMID:29593264)
- Kanda R et al. Expression of the glucagon-like peptide-1 receptor and its role in regulating autophagy in endometrial cancer. BMC Cancer 18, 657 (2018)(PMID:29907137)
- Vrahnas C et al. Author Correction: Increased autophagy in EphrinB2-deficient osteocytes is associated with elevated secondary mineralization and brittle bone. Nat Commun. 10, 5073 (2019)(PMID:31685818)
- Du TT et al. Anterior thalamic nucleus stimulation protects hippocampal neurons by activating autophagy in epileptic monkey. Aging (Albany NY). 12, 6324-6339 (2020)(PMID:32267832)
- Rahaman MS et al. Curcumin alleviates arsenic-induced toxicity in PC12 cells via modulating autophagy/apoptosis. Ecotoxicol Environ Saf. 200, 110756 (2020)(PMID:32464442)
- Rahaman MS et al. Effects of curcumin, D-pinitol alone or in combination in cytotoxicity induced by arsenic in PC12 cells. Food Chem Toxicol. 144, 111577 (2020)(PMID:32679288)
Immunoprecipitation - Hou B et al. SQSTM1/p62 loss reverses the inhibitory effect of sunitinib on autophagy independent of AMPK signaling. Sci Rep. 9, 1087 (2019)(PMID:31366950)
Flow Cytometry - Eng KE et al. A novel quantitative flow cytometry-based assay for autophagy. Autophagy. 6, 634-41 (2010)(PMID:20458170)
Immunocytochemistry - Thayanidhi N et al. Alpha-synuclein delays endoplasmic reticulum (ER)-to-Golgi transport in mammalian cells by antagonizing ER/Golgi SNAREs. Mol Biol Cell. 21, 1850-63 (2010)(PMID:20392839)
- Kovacs JR et al. Autophagy promotes T-cell survival through degradation of proteins of the cell death machinery. Cell Death Differ. 19, 144-52 (2012)(PMID:21660048)
- Cai T et al. Deletion of Ia-2 and/or Ia-2β in mice decreases insulin secretion by reducing the number of dense core vesicles. Diabetologia. 54, 2347-57 (2011)(PMID:21732083)
- Traver MK et al. Immunity-related GTPase M (IRGM) proteins influence the localization of guanylate-binding protein 2 (GBP2) by modulating macroautophagy. J Biol Chem. 286, 30471-80 (2011)(PMID:21757726)
- Moreau K et al. Autophagosome precursor maturation requires homotypic fusion. Cell. 146, 303-17 (2011)(PMID:21784250)
- Gillis JM et al. Aminopeptidase-resistant peptides are targeted to lysosomes and subsequently degraded. Traffic. 12, 1897-910 (2011)(PMID:21883763)
- Seto S et al. Coronin-1a inhibits autophagosome formation around Mycobacterium tuberculosis-containing phagosomes and assists mycobacterial survival in macrophages. Cell Microbiol. 14, 710-27 (2012)(PMID:22256790)
- Starr T et al. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe. 11, 33-45 (2012)(PMID:22264511)
- Sims JJ et al. Polyubiquitin-sensor proteins reveal localization and linkage-type dependence of cellular ubiquitin signaling. Nat Methods. 9, 303-9 (2012)(PMID:22306808)
- McKnight NC et al. Genome-wide siRNA screen reveals amino acid starvation-induced autophagy requires SCOC and WAC. EMBO J. 31, 1931-46 (2012)(PMID:22354037)
- Morris CR et al. A knockout of the Tsg101 gene leads to decreased expression of ErbB receptor tyrosine kinases and induction of autophagy prior to cell death. PLoS One. 7, e34308 (2012)(PMID:22479596)
- Matsuzawa Y et al. TNFAIP3 promotes survival of CD4 T cells by restricting MTOR and promoting autophagy. Autophagy 11, 1052-62 (2015)(PMID:26043155)
- Silva BJ et al. Autophagy Is an Innate Mechanism Associated with Leprosy Polarization. PLoS Pathog. 13, e1006103 (2017)(PMID:28056107)
- Abe S et al. Localization of Protein Kinase NDR2 to Peroxisomes and Its Role in Ciliogenesis. J Biol Chem. 292, 4089-4098 (2017)(PMID:28122914)
- Hasegawa H et al. Single amino acid substitution in LC-CDR1 induces Russell body phenotype that attenuates cellular protein synthesis through eIF2α phosphorylation and thereby downregulates IgG secretion despite operational secretory pathway traffic. MAbs. 9, 854-873 (2017)(PMID:28379093)
- Arasaki K et al. Legionella effector Lpg1137 shuts down ER-mitochondria communication through cleavage of syntaxin 17. Nat Commun. 8, 15406 (2017)(PMID:28504273)
- Wacker R et al. LC3-association with the parasitophorous vacuole membrane of Plasmodium berghei liver stages follows a noncanonical autophagy pathway. Cell Microbiol. 19, e12754 (2017)(PMID:28573684)
- Zhao Q et al. Dual Roles of Two Isoforms of Autophagy-related Gene ATG10 in HCV-Subgenomic replicon Mediated Autophagy Flux and Innate Immunity. Sci Rep. 7, 11250 (2017)(PMID:28900156)
- Newton T et al. Mechanistic basis of an epistatic interaction reducing age at onset in hereditary spastic paraplegia. Brain. 141, 1286-1299 (2018)(PMID:29481671)
- Konno H et al. Pro-inflammation Associated with a Gain-of-Function Mutation (R284S) in the Innate Immune Sensor STING. Cell Rep. 23, 1112-1123 (2018)(PMID:29694889)
- Wise JP Jr et al. Autophagy Disruptions Associated With Altered Optineurin Expression in Extranigral Regions in a Rotenone Model of Parkinson's Disease. Front Neurosci. 12, 289 (2018)(PMID:29867311)
- Kanda R et al. Expression of the glucagon-like peptide-1 receptor and its role in regulating autophagy in endometrial cancer. BMC Cancer 18, 657 (2018)(PMID:29907137)
- Mori H et al. Induction of selective autophagy in cells replicating hepatitis C virus genome. J Gen Virol. 99, 1643-1657 (2018)(PMID:30311874)
- Kajiume T, Kobayashi M, Human granulocytes undergo cell death via autophagy. Cell Death Discov. 4, 111 (2018)
(PMID:30534419)
- Bordi M et al. mTOR hyperactivation in Down Syndrome underlies deficits in autophagy induction, autophagosome formation, and mitophagy. Cell Death Dis. 10, 563 (2019)(PMID:31332166)
- Lasič E et al. Astrocyte Specific Remodeling of Plasmalemmal Cholesterol Composition by Ketamine Indicates a New Mechanism of Antidepressant Action. Sci Rep. 9, 10957 (2019)(PMID:31358895)
- Vrahnas C et al. Author Correction: Increased autophagy in EphrinB2-deficient osteocytes is associated with elevated secondary mineralization and brittle bone. Nat Commun. 10, 5073 (2019)(PMID:31685818)
- Ye X et al. Lipopolysaccharide Induces Neuroinflammation in Microglia by Activating the MTOR Pathway and Downregulating Vps34 to Inhibit Autophagosome Formation. J Neuroinflammation. 17, 18 (2020)(PMID:31926553)
- Boukhalfa A et al. PI3KC2α-dependent and VPS34-independent Generation of PI3P Controls Primary Cilium-Mediated Autophagy in Response to Shear Stress. Nat Commun. 11, 294 (2020)(PMID:31941925)
Immunohistochemistry - Chalazonitis A et al. Homeodomain interacting protein kinase 2 regulates postnatal development of enteric dopaminergic neurons and glia via BMP signaling. J Neurosci. 31, 13746-57 (2011)(PMID:21957238)
- Silva BJ et al. Autophagy Is an Innate Mechanism Associated with Leprosy Polarization. PLoS Pathog. 13, e1006103 (2017)(PMID:28056107)
- Nakano S et al. The role of p62/SQSTM1 in sporadic inclusion body myositis. Neuromuscul Disord. 27, 363-369 (2017)(PMID:28159418)
- Kimura T et al. Age-dependent changes in synaptic plasticity enhance tau oligomerization in the mouse hippocampus. Acta Neuropathol Commun. 5, 67 (2017)(PMID:28874186)
- Sato Y et al. Apobec2 deficiency causes mitochondrial defects and mitophagy in skeletal muscle. FASEB J. 32, 1428-1439 (2018)(PMID:29127187)
Immunofluorescence - Zemirli N et al. The primary cilium protein folliculin is part of the autophagy signaling pathway to regulate epithelial cell size in response to fluid flow. Cell Stress. 3, 100-109 (2019)(PMID:31225504)
Other(Immuno-EM) - Murthy A et al. A Crohn's disease variant in Atg16l1 enhances its degradation by caspase 3. Nature 506, 456-62 (2014)(PMID:24553140)
Other(Image-based flow cytometry) - Murthy A et al. A Crohn's disease variant in Atg16l1 enhances its degradation by caspase 3. Nature 506, 456-62 (2014) (PMID:24553140)
- Perot BP et al. Autophagy diminishes the early interferon-β response to influenza A virus resulting in differential expression of interferon-stimulated genes. Cell Death Dis. 9, 539 (2018)(PMID:29748576)
|
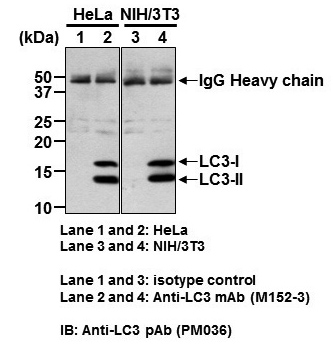
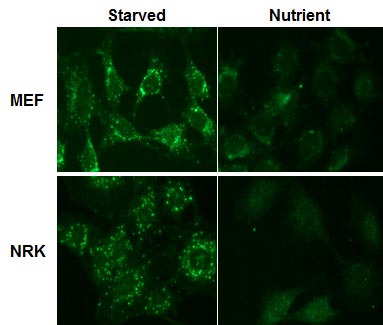

 Copyright (C)2015 Medical & Biological Laboratories Co., Ltd. All Rights Reserve
Copyright (C)2015 Medical & Biological Laboratories Co., Ltd. All Rights Reserve